{
"cells": [
{
"cell_type": "markdown",
"id": "73521bac",
"metadata": {},
"source": [
"# 3. Generating a 4D-Communication Tensor from computed communication scores\n",
"\n",
"After inferring communication scores for combinations of ligand-receptor and sender-receiver cell pairs, we can use that information to identify context-dependent CCC patterns across multiple samples simultaneously by generating a 4D-Communication Tensor. LIANA handily outputs these score as a dataframe that is easy to use for building our tensor.\n",
"\n",
"In this tutorial we will show you how to use the dataframe saved from LIANA to generate a 4D-Communication Tensor that could be later used with Tensor-cell2cell."
]
},
{
"cell_type": "markdown",
"id": "739c7ec9",
"metadata": {},
"source": [
"## Initial Setup\n",
"\n",
"**Before running this notebook** \n",
"\n",
"GPUs can substantially speed-up the downstream decomposition. If you are planning to use a NVIDIA GPU, make sure to have a proper NVIDIA GPU driver (https://www.nvidia.com/Download/index.aspx) as well as the CUDA toolkit (https://developer.nvidia.com/cuda-toolkit) installed.\n",
"\n",
"Then, make sure to create an environment with PyTorch version >= v1.8.1 following these instructions to enable CUDA.\n",
"\n",
"https://pytorch.org/get-started/locally/\n",
"\n",
"### Enabling GPU use\n",
"\n",
"If you are using a NVIDIA GPU, after installing PyTorch with CUDA enabled, specify `gpu_use = True`. Otherwise, `gpu_use = False`. In R, this must be specified during tensor building. "
]
},
{
"cell_type": "code",
"execution_count": 1,
"id": "88f2b541",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [],
"source": [
"library(reticulate, quietly = TRUE)\n",
"\n",
"gpu_use = TRUE\n",
"if (gpu_use){\n",
" device <- 'cuda:0'\n",
" tensorly <- reticulate::import('tensorly')\n",
" tensorly$set_backend('pytorch')\n",
"}else{\n",
" device <- NULL\n",
"}"
]
},
{
"cell_type": "markdown",
"id": "9cefd77f",
"metadata": {},
"source": [
"**load the necessary libraries**"
]
},
{
"cell_type": "code",
"execution_count": 2,
"id": "50007cc4",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [],
"source": [
"library(liana, quietly = TRUE)"
]
},
{
"cell_type": "code",
"execution_count": 3,
"id": "9b58b2c9",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [],
"source": [
"data_folder = '../../data/liana-outputs/'\n",
"output_folder = '../../data/tc2c-outputs/'\n",
"\n",
"env_name = 'ccc_protocols' # conda environment created with ../../env_setup/setup_env.sh"
]
},
{
"cell_type": "markdown",
"id": "c7e02ce4",
"metadata": {},
"source": [
"## Load Data\n",
"\n",
"Open the list containing LIANA results for each sample/context. These results contain the communication scores of the combinations of ligand-receptor pairs and sender-receiver pairs."
]
},
{
"cell_type": "code",
"execution_count": 4,
"id": "58d5fd23",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [],
"source": [
"context_df_dict <- readRDS(file.path(data_folder, 'LIANA_by_sample_R.rds'))"
]
},
{
"cell_type": "markdown",
"id": "81e96bd1",
"metadata": {},
"source": [
"## Create 4D-Communication Tensor\n",
"\n",
"### Specify the order of the samples/contexts\n",
"\n",
"Here, we will specify an order of the samples/contexts given the condition they belong to (HC or *Control*, M or *Moderate COVID-19*, S or *Severe COVID-19*)."
]
},
{
"cell_type": "code",
"execution_count": 5,
"id": "d89ce3fa",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [],
"source": [
"sorted_samples = sort(names(context_df_dict))"
]
},
{
"cell_type": "code",
"execution_count": 6,
"id": "dcff0d18",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [
{
"data": {
"text/html": [
"\n",
"- 'HC1'
- 'HC2'
- 'HC3'
- 'M1'
- 'M2'
- 'M3'
- 'S1'
- 'S2'
- 'S3'
- 'S4'
- 'S5'
- 'S6'
\n"
],
"text/latex": [
"\\begin{enumerate*}\n",
"\\item 'HC1'\n",
"\\item 'HC2'\n",
"\\item 'HC3'\n",
"\\item 'M1'\n",
"\\item 'M2'\n",
"\\item 'M3'\n",
"\\item 'S1'\n",
"\\item 'S2'\n",
"\\item 'S3'\n",
"\\item 'S4'\n",
"\\item 'S5'\n",
"\\item 'S6'\n",
"\\end{enumerate*}\n"
],
"text/markdown": [
"1. 'HC1'\n",
"2. 'HC2'\n",
"3. 'HC3'\n",
"4. 'M1'\n",
"5. 'M2'\n",
"6. 'M3'\n",
"7. 'S1'\n",
"8. 'S2'\n",
"9. 'S3'\n",
"10. 'S4'\n",
"11. 'S5'\n",
"12. 'S6'\n",
"\n",
"\n"
],
"text/plain": [
" [1] \"HC1\" \"HC2\" \"HC3\" \"M1\" \"M2\" \"M3\" \"S1\" \"S2\" \"S3\" \"S4\" \"S5\" \"S6\" "
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"sorted_samples"
]
},
{
"cell_type": "code",
"execution_count": 7,
"id": "bd3d191e",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [
{
"data": {
"text/html": [
"\n",
"A tibble: 6 × 15\n",
"\n",
"\t| source | target | ligand.complex | receptor.complex | magnitude_rank | specificity_rank | magnitude_mean_rank | natmi.prod_weight | sca.LRscore | cellphonedb.lr.mean | specificity_mean_rank | natmi.edge_specificity | connectome.weight_sc | logfc.logfc_comb | cellphonedb.pvalue |
|---|
\n",
"\t| <chr> | <chr> | <chr> | <chr> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> |
|---|
\n",
"\n",
"\n",
"\t| T | NK | B2M | CD3D | 3.983438e-11 | 0.0134542495 | 1.000000 | 8.059861 | 0.9610254 | 3.410586 | 487.75 | 0.08327606 | 1.3008562 | 0.8719799 | 0 |
\n",
"\t| Macrophages | NK | B2M | CD3D | 3.186750e-10 | 0.0135072450 | 2.000000 | 8.059599 | 0.9610248 | 3.410500 | 487.00 | 0.08327335 | 1.3005565 | 0.8832177 | 0 |
\n",
"\t| NK | NK | B2M | CD3D | 4.979297e-09 | 0.0181475450 | 3.666667 | 7.614378 | 0.9599465 | 3.264099 | 571.25 | 0.07867324 | 0.7909129 | 0.6978778 | 0 |
\n",
"\t| T | NK | B2M | KLRD1 | 1.366319e-08 | 0.0003588748 | 5.666667 | 6.865250 | 0.9579074 | 3.297900 | 211.75 | 0.17129308 | 6.9609202 | 0.8624079 | 0 |
\n",
"\t| B | NK | B2M | CD3D | 2.039520e-08 | 0.0192306877 | 5.333333 | 7.518543 | 0.9597023 | 3.232586 | 595.00 | 0.07768305 | 0.6812107 | 0.6910179 | 0 |
\n",
"\t| Macrophages | NK | B2M | KLRD1 | 2.039520e-08 | 0.0003588748 | 6.666667 | 6.865027 | 0.9579067 | 3.297814 | 210.75 | 0.17128751 | 6.9606205 | 0.8736456 | 0 |
\n",
"\n",
"
\n"
],
"text/latex": [
"A tibble: 6 × 15\n",
"\\begin{tabular}{lllllllllllllll}\n",
" source & target & ligand.complex & receptor.complex & magnitude\\_rank & specificity\\_rank & magnitude\\_mean\\_rank & natmi.prod\\_weight & sca.LRscore & cellphonedb.lr.mean & specificity\\_mean\\_rank & natmi.edge\\_specificity & connectome.weight\\_sc & logfc.logfc\\_comb & cellphonedb.pvalue\\\\\n",
" & & & & & & & & & & & & & & \\\\\n",
"\\hline\n",
"\t T & NK & B2M & CD3D & 3.983438e-11 & 0.0134542495 & 1.000000 & 8.059861 & 0.9610254 & 3.410586 & 487.75 & 0.08327606 & 1.3008562 & 0.8719799 & 0\\\\\n",
"\t Macrophages & NK & B2M & CD3D & 3.186750e-10 & 0.0135072450 & 2.000000 & 8.059599 & 0.9610248 & 3.410500 & 487.00 & 0.08327335 & 1.3005565 & 0.8832177 & 0\\\\\n",
"\t NK & NK & B2M & CD3D & 4.979297e-09 & 0.0181475450 & 3.666667 & 7.614378 & 0.9599465 & 3.264099 & 571.25 & 0.07867324 & 0.7909129 & 0.6978778 & 0\\\\\n",
"\t T & NK & B2M & KLRD1 & 1.366319e-08 & 0.0003588748 & 5.666667 & 6.865250 & 0.9579074 & 3.297900 & 211.75 & 0.17129308 & 6.9609202 & 0.8624079 & 0\\\\\n",
"\t B & NK & B2M & CD3D & 2.039520e-08 & 0.0192306877 & 5.333333 & 7.518543 & 0.9597023 & 3.232586 & 595.00 & 0.07768305 & 0.6812107 & 0.6910179 & 0\\\\\n",
"\t Macrophages & NK & B2M & KLRD1 & 2.039520e-08 & 0.0003588748 & 6.666667 & 6.865027 & 0.9579067 & 3.297814 & 210.75 & 0.17128751 & 6.9606205 & 0.8736456 & 0\\\\\n",
"\\end{tabular}\n"
],
"text/markdown": [
"\n",
"A tibble: 6 × 15\n",
"\n",
"| source <chr> | target <chr> | ligand.complex <chr> | receptor.complex <chr> | magnitude_rank <dbl> | specificity_rank <dbl> | magnitude_mean_rank <dbl> | natmi.prod_weight <dbl> | sca.LRscore <dbl> | cellphonedb.lr.mean <dbl> | specificity_mean_rank <dbl> | natmi.edge_specificity <dbl> | connectome.weight_sc <dbl> | logfc.logfc_comb <dbl> | cellphonedb.pvalue <dbl> |\n",
"|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|\n",
"| T | NK | B2M | CD3D | 3.983438e-11 | 0.0134542495 | 1.000000 | 8.059861 | 0.9610254 | 3.410586 | 487.75 | 0.08327606 | 1.3008562 | 0.8719799 | 0 |\n",
"| Macrophages | NK | B2M | CD3D | 3.186750e-10 | 0.0135072450 | 2.000000 | 8.059599 | 0.9610248 | 3.410500 | 487.00 | 0.08327335 | 1.3005565 | 0.8832177 | 0 |\n",
"| NK | NK | B2M | CD3D | 4.979297e-09 | 0.0181475450 | 3.666667 | 7.614378 | 0.9599465 | 3.264099 | 571.25 | 0.07867324 | 0.7909129 | 0.6978778 | 0 |\n",
"| T | NK | B2M | KLRD1 | 1.366319e-08 | 0.0003588748 | 5.666667 | 6.865250 | 0.9579074 | 3.297900 | 211.75 | 0.17129308 | 6.9609202 | 0.8624079 | 0 |\n",
"| B | NK | B2M | CD3D | 2.039520e-08 | 0.0192306877 | 5.333333 | 7.518543 | 0.9597023 | 3.232586 | 595.00 | 0.07768305 | 0.6812107 | 0.6910179 | 0 |\n",
"| Macrophages | NK | B2M | KLRD1 | 2.039520e-08 | 0.0003588748 | 6.666667 | 6.865027 | 0.9579067 | 3.297814 | 210.75 | 0.17128751 | 6.9606205 | 0.8736456 | 0 |\n",
"\n"
],
"text/plain": [
" source target ligand.complex receptor.complex magnitude_rank\n",
"1 T NK B2M CD3D 3.983438e-11 \n",
"2 Macrophages NK B2M CD3D 3.186750e-10 \n",
"3 NK NK B2M CD3D 4.979297e-09 \n",
"4 T NK B2M KLRD1 1.366319e-08 \n",
"5 B NK B2M CD3D 2.039520e-08 \n",
"6 Macrophages NK B2M KLRD1 2.039520e-08 \n",
" specificity_rank magnitude_mean_rank natmi.prod_weight sca.LRscore\n",
"1 0.0134542495 1.000000 8.059861 0.9610254 \n",
"2 0.0135072450 2.000000 8.059599 0.9610248 \n",
"3 0.0181475450 3.666667 7.614378 0.9599465 \n",
"4 0.0003588748 5.666667 6.865250 0.9579074 \n",
"5 0.0192306877 5.333333 7.518543 0.9597023 \n",
"6 0.0003588748 6.666667 6.865027 0.9579067 \n",
" cellphonedb.lr.mean specificity_mean_rank natmi.edge_specificity\n",
"1 3.410586 487.75 0.08327606 \n",
"2 3.410500 487.00 0.08327335 \n",
"3 3.264099 571.25 0.07867324 \n",
"4 3.297900 211.75 0.17129308 \n",
"5 3.232586 595.00 0.07768305 \n",
"6 3.297814 210.75 0.17128751 \n",
" connectome.weight_sc logfc.logfc_comb cellphonedb.pvalue\n",
"1 1.3008562 0.8719799 0 \n",
"2 1.3005565 0.8832177 0 \n",
"3 0.7909129 0.6978778 0 \n",
"4 6.9609202 0.8624079 0 \n",
"5 0.6812107 0.6910179 0 \n",
"6 6.9606205 0.8736456 0 "
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"head(context_df_dict$HC1)"
]
},
{
"cell_type": "markdown",
"id": "f4a27f73",
"metadata": {},
"source": [
"## Generate tensor\n",
"\n",
"To generate the 4D-communication tensor, we will to create matrices with the communication scores for each of the ligand-receptor pairs within the same sample, then generate a 3D tensor for each sample, and finally concatenate them to form the 4D tensor.\n",
"\n",
"Briefly, we use the LIANA dataframe and communication scores to organize them as follows:\n",
"\n",
"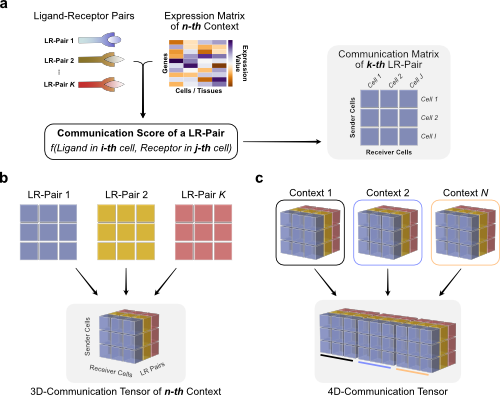"
]
},
{
"cell_type": "markdown",
"id": "6e0e5670",
"metadata": {},
"source": [
"Next we will apply some additional preprocessing (transformations/filtering) to the communication scores. Important parameters include:\n",
"\n",
"- `non_negative` and `non_negative_fill` as Tensor-cell2cell by default expects non-negative values, any values below 0 will be set to 0 (e.g. this is relevant if one wants to use e.g. the `LRlog2FC` score function). If you used a pipeline that generated negative scores, we suggest replacing these with 0. Otherwise, by default, Tensor-cell2cell will treat these as NaN. Since we used the magnitude rank score, which is non-negative, these parameters won't affect our results. \n",
"\n",
"- `invert` and `invert_fun` convert the communication score before using it to build the tensor. In this case, the 'magnitude_rank' score generated by LIANA considers low values as the most important ones, ranging from 0 to 1. In contrast, Tensor-cell2cell requires higher values to be the most important scores, so here we pass a function (`function (x) 1 - x`) to adapt LIANA's magnitude-rank scores (subtracts the LIANA's score from 1). If using other scores coming from one of the methods implemented in LIANA, a similar transformation can be done depending on the parameters and assumptions of the scoring method.\n",
"\n",
"- `outer_frac` controls the elements to include in the union scenario of the `how` options. Only elements that are present at least in this fraction of samples/contexts will be included. When this value is 0, the tensor includes all elements across the samples. When this value is 1, it acts as using `how='inner'`."
]
},
{
"cell_type": "code",
"execution_count": 8,
"id": "51d82570",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [
{
"name": "stderr",
"output_type": "stream",
"text": [
"Inverting `magnitude_rank`!\n",
"\n",
"Inverting `magnitude_rank`!\n",
"\n",
"Inverting `magnitude_rank`!\n",
"\n",
"Inverting `magnitude_rank`!\n",
"\n",
"Inverting `magnitude_rank`!\n",
"\n",
"Inverting `magnitude_rank`!\n",
"\n",
"Inverting `magnitude_rank`!\n",
"\n",
"Inverting `magnitude_rank`!\n",
"\n",
"Inverting `magnitude_rank`!\n",
"\n",
"Inverting `magnitude_rank`!\n",
"\n",
"Inverting `magnitude_rank`!\n",
"\n",
"Inverting `magnitude_rank`!\n",
"\n"
]
}
],
"source": [
"context_df_dict <- liana:::preprocess_scores(context_df_dict = context_df_dict, \n",
" outer_frac = 1/3, # Fraction of samples as threshold to include cells and LR pairs.\n",
" invert = TRUE, # transform the scores\n",
" invert_fun = function(x) 1-x, # Transformation function\n",
" non_negative = TRUE, # fills negative values\n",
" non_negative_fill = 0 # set negative values to 0\n",
" )"
]
},
{
"cell_type": "markdown",
"id": "cd74ee96",
"metadata": {},
"source": [
"Next, we will transform the structure of the communication scores from a set of 2D-matrices for each sample into a 3D Tensor where the third dimension is sample/context. **The key parameters when building a tensor are:**\n",
"\n",
"- `how` controls which ligand-receptor pairs and cell types to include when building the tensor. This decision depends on whether the missing values across a number of samples for both ligand-receptor interactions and sender-receiver cell pairs are considered to be biologically-relevant. Options are:\n",
" - `'inner'` is the more strict option since it only considers only cell types and LR pairs that are present in all contexts (intersection).\n",
" - `'outer'` considers all cell types and LR pairs that are present across contexts (union).\n",
" - `'outer_lrs'` considers only cell types that are present in all contexts (intersection), while all LR pairs that are present across contexts (union).\n",
" - `'outer_cells'` considers only LR pairs that are present in all contexts (intersection), while all cell types that are present across contexts (union).\n",
"\n",
"- `fill` indicates what value to assign missing scores when `how` is set to `'outer'`, i.e., those cells or LR pairs that are not present in all contexts. During tensor component analysis, NaN values are masked such that they are not considered by the decomposition objective. This results in behavior of NaNs being imputed as missing values that are potentially communicating, whereas if missing LRs are filled with a value such as 0, they are treated as biological zeroes (i.e., not communicating). For additional details and discussion regarding this parameter, please see the [missing indices benchmarking](../../tc2c_benchmark/scripts/missing_indices_consistency.ipynb).\n",
" - `'lf_fill'`: value to fill missing (across contexts) LR pairs with\n",
" - `'cell_fill'`: value to fill missing (across contexts OR LR pairs) cell types with"
]
},
{
"cell_type": "code",
"execution_count": 9,
"id": "f2217458",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"[1] 0\n"
]
},
{
"name": "stderr",
"output_type": "stream",
"text": [
"Loading `ccc_protocols` Conda Environment\n",
"\n",
"Building the tensor using magnitude_rank...\n",
"\n"
]
}
],
"source": [
"tensor <- liana_tensor_c2c(context_df_dict = context_df_dict,\n",
" sender_col = \"source\", # Column name of the sender cells\n",
" receiver_col = \"target\", # Column name of the receiver cells\n",
" ligand_col = \"ligand.complex\", # Column name of the ligands\n",
" receptor_col = \"receptor.complex\", # Column name of the receptors\n",
" score_col = 'magnitude_rank', # Column name of the communication scores to use\n",
" how='outer', # What to include across all samples\n",
" lr_fill = NaN, # What to fill missing LRs with \n",
" cell_fill = NaN, # What to fill missing cell types with \n",
" lr_sep='^', # How to separate ligand and receptor names to name LR pair\n",
" context_order=sorted_samples, # Order to store the contexts in the tensor\n",
" sort_elements = TRUE, # Whether sorting alphabetically element names of each tensor dim. Does not apply for context order if context_order is passed.\n",
" conda_env = 'ccc_protocols', # used to pass an existing conda env with cell2cell\n",
" build_only = TRUE, , # set this to FALSE to combine the downstream rank selection and decomposition steps all here \n",
" device = device # Device to use when backend is pytorch.\n",
" )"
]
},
{
"cell_type": "markdown",
"id": "745d7311",
"metadata": {},
"source": [
"## Evaluate some tensor properties\n",
"\n",
"### Tensor shape\n",
"This indicates the number of elements in each tensor dimension: (Contexts, LR pairs, Sender cells, Receiver cells)"
]
},
{
"cell_type": "code",
"execution_count": 10,
"id": "bfa209d6",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [
{
"data": {
"text/plain": [
"torch.Size([12, 1054, 10, 10])"
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"tensor$shape"
]
},
{
"cell_type": "markdown",
"id": "8b8038ce",
"metadata": {},
"source": [
"### Missing values\n",
"This represents the fraction of values that are missing. In this case, missing values are combinations of contexts x LR pairs x Sender cells x Receiver cells that did not have a communication score or were missing in the dataframes."
]
},
{
"cell_type": "code",
"execution_count": 11,
"id": "3b8df693",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [
{
"data": {
"text/html": [
"0.906289577484131"
],
"text/latex": [
"0.906289577484131"
],
"text/markdown": [
"0.906289577484131"
],
"text/plain": [
"[1] 0.9062896"
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"tensor$missing_fraction()"
]
},
{
"cell_type": "markdown",
"id": "021b564d",
"metadata": {},
"source": [
"### Sparsity\n",
"This represents the fraction of values that are a real zero (excluding the missing values)"
]
},
{
"cell_type": "code",
"execution_count": 12,
"id": "4abcc690",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [
{
"data": {
"text/html": [
"0.0307012982666492"
],
"text/latex": [
"0.0307012982666492"
],
"text/markdown": [
"0.0307012982666492"
],
"text/plain": [
"[1] 0.0307013"
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"tensor$sparsity_fraction()"
]
},
{
"cell_type": "markdown",
"id": "b1a3c7de",
"metadata": {},
"source": [
"### Fraction of excluded elements\n",
"This represents the fraction of values that are ignored (masked) in the analysis. In this case it coincides with the missing values because we did not generate a new `tensor.mask` to manually ignore specific values. Instead, it automatically excluded the missing values."
]
},
{
"cell_type": "code",
"execution_count": 13,
"id": "3aed0f75",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [
{
"data": {
"text/html": [
"0.906289525330067"
],
"text/latex": [
"0.906289525330067"
],
"text/markdown": [
"0.906289525330067"
],
"text/plain": [
"[1] 0.9062895"
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"tensor$excluded_value_fraction() # Percentage of values in the tensor that are masked/missing"
]
},
{
"cell_type": "markdown",
"id": "def0d439",
"metadata": {},
"source": [
"## Export Tensor\n",
"\n",
"Here we will save the `tensor` so we can use it later with other analyses."
]
},
{
"cell_type": "code",
"execution_count": 14,
"id": "bbc76f90",
"metadata": {
"vscode": {
"languageId": "r"
}
},
"outputs": [],
"source": [
"reticulate::py_save_object(object = tensor, \n",
" filename = file.path(output_folder, 'BALF-Tensor-R.pkl'))"
]
}
],
"metadata": {
"kernelspec": {
"display_name": "R [conda env:ccc_protocols]",
"language": "R",
"name": "conda-env-ccc_protocols-r"
},
"language_info": {
"codemirror_mode": "r",
"file_extension": ".r",
"mimetype": "text/x-r-source",
"name": "R",
"pygments_lexer": "r",
"version": "4.2.3"
}
},
"nbformat": 4,
"nbformat_minor": 5
}