S3B. Decomposition Consistency between Score Types
We can also check how consistent each score type is after running the full Tensor-cell2cell pipeline. The CorrIndex provides a dissimilarity metric that can compare decompositions of the same rank.
[1]:
from collections import defaultdict
import itertools
import os
import matplotlib
import numpy as np
import pandas as pd
import torch
import liana as li
import cell2cell as c2c
from cell2cell.tensor.metrics import correlation_index
[2]:
if torch.cuda.is_available():
import tensorly as tl
tl.set_backend('pytorch')
device = 'cuda:0'
else:
device = 'cpu'
[3]:
data_folder = '../../data/liana-outputs/'
output_folder = '../../data/tc2c-outputs/'
c2c.io.directories.create_directory(output_folder)
../../data/tc2c-outputs/ already exists.
First, let’s load the LIANA scores for each sample and score type:
[4]:
liana_res = pd.read_csv(data_folder + 'LIANA_by_sample.csv')
sorted_samples = sorted(liana_res['sample_new'].unique())
Next, we can generate and decompose the tensor as in Tutorial 03 and Tutorial 04 respectively, but for each magnitude rank score separately. We will use the rank identified by the consensus magnitude score in Tutorial 04.
[5]:
tf_optimization_dict = {'regular': {'runs': 1,
'tol': 10e-7,
'n_iter_max': 100},
'robust': {'runs': 100,
'tol': 10e-8,
'n_iter_max': 500}}
def run_tensor_pipeline(liana_res, score_type, rank,
tf_optimization = 'robust'):
# build tensor
tensor = li.multi.to_tensor_c2c(liana_res=liana_res, # LIANA's dataframe containing results
sample_key='sample_new', # Column name of the samples
source_key='source', # Column name of the sender cells
target_key='target', # Column name of the receiver cells
ligand_key='ligand_complex', # Column name of the ligands
receptor_key='receptor_complex', # Column name of the receptors
score_key=score_type, # Column name of the communication scores to use
non_negative = True, # set negative values to 0
inverse_fun=lambda x: x, # Transformation function -- don't invert because it's not a rank score
non_expressed_fill=None, # Value to replace missing values with
how='outer', # What to include across all samples
lr_fill=np.nan, # What to fill missing LRs with
cell_fill = np.nan, # What to fill missing cell types with
outer_fraction=1/3., # Fraction of samples as threshold to include cells and LR pairs.
lr_sep='^', # How to separate ligand and receptor names to name LR pair
context_order=sorted_samples, # Order to store the contexts in the tensor
sort_elements=True # Whether sorting alphabetically element names of each tensor dim. Does not apply for context order if context_order is passed.
)
# get metadata
element_dict = defaultdict(lambda: 'Unknown')
context_dict = element_dict.copy()
context_dict.update({'HC1' : 'Control',
'HC2' : 'Control',
'HC3' : 'Control',
'M1' : 'Moderate COVID-19',
'M2' : 'Moderate COVID-19',
'M3' : 'Moderate COVID-19',
'S1' : 'Severe COVID-19',
'S2' : 'Severe COVID-19',
'S3' : 'Severe COVID-19',
'S4' : 'Severe COVID-19',
'S5' : 'Severe COVID-19',
'S6' : 'Severe COVID-19',
})
dimensions_dict = [context_dict, None, None, None]
meta_tensor = c2c.tensor.generate_tensor_metadata(interaction_tensor=tensor,
metadata_dicts=[context_dict, None, None, None],
fill_with_order_elements=True
)
# decompose tensor
tensor.to_device(device)
tensor.compute_tensor_factorization(rank=rank,
init='random', # Initialization method of the tensor factorization
svd='numpy_svd', # Type of SVD to use if the initialization is 'svd'
random_state=0, # Random seed for reproducibility
normalize_loadings=True,
runs=tf_optimization_dict[tf_optimization]['runs'],
tol=tf_optimization_dict[tf_optimization]['tol'],
n_iter_max=tf_optimization_dict[tf_optimization]['n_iter_max']
)
return tensor
[6]:
tensor_magnitude = c2c.io.read_data.load_tensor(os.path.join(output_folder, 'BALF-Tensor_decomposed.pkl'))
magnitude_scores = ['lr_means', 'expr_prod', 'lrscore', 'lr_probs']
/home/hratch/miniconda3/envs/ccc_protocols/lib/python3.10/site-packages/tensorly/backend/pytorch_backend.py:39: UserWarning: To copy construct from a tensor, it is recommended to use sourceTensor.clone().detach() or sourceTensor.clone().detach().requires_grad_(True), rather than torch.tensor(sourceTensor).
[7]:
tensors_individual = {}
# calculate on each magnitude score type separately
for score_type in magnitude_scores:
ti = run_tensor_pipeline(liana_res = liana_res,
score_type = score_type,
rank = tensor_magnitude.rank,
tf_optimization = 'robust')
ti.to_device('cpu') # ensures can be pickled properly
tensors_individual[score_type] = ti
c2c.io.export_variable_with_pickle(tensors_individual, output_folder + '/BALF-Tensor_individualscores.pkl')
tensors_individual = c2c.io.read_data.load_variable_with_pickle(output_folder + '/BALF-Tensor_individualscores.pkl')
100%|███████████████████████████████████████████| 12/12 [00:18<00:00, 1.54s/it]
/home/hratch/miniconda3/envs/ccc_protocols/lib/python3.10/site-packages/tensorly/backend/pytorch_backend.py:39: UserWarning: To copy construct from a tensor, it is recommended to use sourceTensor.clone().detach() or sourceTensor.clone().detach().requires_grad_(True), rather than torch.tensor(sourceTensor).
100%|█████████████████████████████████████████| 100/100 [05:50<00:00, 3.50s/it]
Best model has a normalized error of: 0.207
100%|███████████████████████████████████████████| 12/12 [00:18<00:00, 1.54s/it]
/home/hratch/miniconda3/envs/ccc_protocols/lib/python3.10/site-packages/tensorly/backend/pytorch_backend.py:39: UserWarning: To copy construct from a tensor, it is recommended to use sourceTensor.clone().detach() or sourceTensor.clone().detach().requires_grad_(True), rather than torch.tensor(sourceTensor).
100%|█████████████████████████████████████████| 100/100 [05:46<00:00, 3.46s/it]
Best model has a normalized error of: 0.340
100%|███████████████████████████████████████████| 12/12 [00:18<00:00, 1.54s/it]
/home/hratch/miniconda3/envs/ccc_protocols/lib/python3.10/site-packages/tensorly/backend/pytorch_backend.py:39: UserWarning: To copy construct from a tensor, it is recommended to use sourceTensor.clone().detach() or sourceTensor.clone().detach().requires_grad_(True), rather than torch.tensor(sourceTensor).
100%|█████████████████████████████████████████| 100/100 [05:31<00:00, 3.31s/it]
Best model has a normalized error of: 0.079
100%|███████████████████████████████████████████| 12/12 [00:18<00:00, 1.54s/it]
/home/hratch/miniconda3/envs/ccc_protocols/lib/python3.10/site-packages/tensorly/backend/pytorch_backend.py:39: UserWarning: To copy construct from a tensor, it is recommended to use sourceTensor.clone().detach() or sourceTensor.clone().detach().requires_grad_(True), rather than torch.tensor(sourceTensor).
100%|█████████████████████████████████████████| 100/100 [05:54<00:00, 3.55s/it]
Best model has a normalized error of: 0.502
../../data/tc2c-outputs//BALF-Tensor_individualscores.pkl was correctly saved.
Next, we can calculate the similarity between each score type’s deomposition output based on the CorrIndex metric. For more information on the CorrIndex, as well as similar analyses on decomposition consistency, see Figure 3A of this paper:
[8]:
corrindex_res = pd.DataFrame(columns = ['score_type_1', 'score_type_2', 'Similarity'])
# calculate corrindex pairwise between score types
for idx, score_type_comparison in enumerate(itertools.permutations(tensors_individual, 2)):
tensor_1 = tensors_individual[score_type_comparison[0]]
tensor_2 = tensors_individual[score_type_comparison[1]]
corrindex = correlation_index(tensor_1.factors, tensor_2.factors)
similarity = 1 - corrindex
corrindex_res.loc[idx, :] = list(score_type_comparison) + [similarity]
# formatting
corrindex_res = corrindex_res.pivot(index='score_type_1', columns='score_type_2', values = 'Similarity')
corrindex_res.columns = corrindex_res.columns.tolist()
corrindex_res.index = corrindex_res.index.tolist()
np.fill_diagonal(corrindex_res.values, 1)
for col_name in corrindex_res.columns:
corrindex_res[col_name] = corrindex_res[col_name].astype(float)
method_name_map = {'expr_prod': 'Connectome/NATMI',
'lr_means': 'CellPhoneDB',
'lr_probs': 'CellChat',
'lrscore': 'SingleCellSignalR'}
corrindex_res.columns = corrindex_res.columns.map(method_name_map).tolist()
corrindex_res.index = corrindex_res.index.map(method_name_map).tolist()
[9]:
cm = c2c.plotting.clustermap_cci(corrindex_res,
method='ward',
optimal_leaf=True,
metadata=None,
title='',
cbar_title='Similarity',
cmap='Blues_r',
vmax=1.,
# vmin=0.,
annot=True,
dendrogram_ratio=0.15,
figsize=(4,5))
font = matplotlib.font_manager.FontProperties(weight='bold', size=7)
for ax in [cm.ax_heatmap, cm.ax_cbar]:
for tick in ax.get_xticklabels():
tick.set_fontproperties(font)
for tick in ax.get_yticklabels():
tick.set_fontproperties(font)
text = ax.yaxis.label
text.set_font_properties(font)
Interaction space detected as a distance matrix
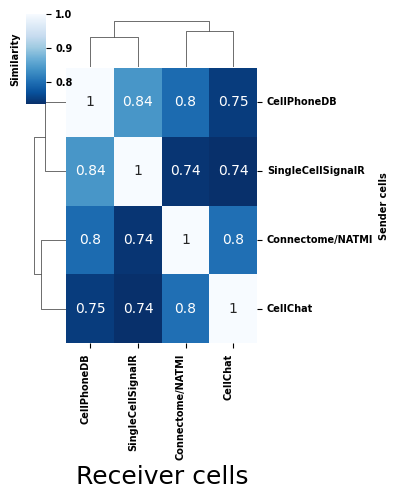
Given the high overall similarity between score types, we see that Tensor-cell2cell’s decomposition tends to capture consistent patterns across samples, smoothing over some of the inconsistencies in communication scores that we saw at the individual sample level in Tutorial 02